AI, Nano, Consciousness, and Eclectic Interests


Hi, I'm Jackson.
I make and am fascinated by nanoscale systems and materials. I believe that some of the world's biggest leaps forward will come from advances at the very small scale, in science and engineering.
Hand in hand with my interest in nanoscale systems is my interest in artificial intelligence and optical devices. My research at Penn State University involved the study, simulation, and fabrication of photodetectors. I worked to find ways to increase the efficiency of photodetectors over a broad range of electromagnetic wavelengths with the help of advanced simulation packages such as COMSOL and Ansys HFSS, using biologically inspired optimization algorithms such as evolutionary programming and artificial neural networks. I also worked in research building machine learning systems to classify output from novel biological devices that interface with the human body.
I have an interdisciplinary background, with professional training in medical physiology, neuroanatomy, neurobiology, and biochemistry - as well as laboratory experience developing and characterizing semiconductor-based devices and nanoparticles. I believe that it is because of my interdisciplinary background that I am able to think differently about engineering solutions, understanding and applying valuable discoveries that researchers have made across multiple fields of study.
I am also an avid programmer and enjoy making visualizations, as well as simulation and optimization software, in my spare time.
Please feel free to see view some of my work by scrolling down:
Portfolio:
Right: This is the result of a project that I did, where I combined genetic algorithms with neural networks in an effort to train a neural network to balance a cartpole (the training environment was provided by OpenAI's gym library). The training took a far shorter amount of time than I expected, and it was a good proof of concept behind the idea of using genetic algorithms for reinforcement learning in neural networks, as opposed to gradient descent techniques. This concept, however, would be even more useful for environments where it is very challenging to define success from moment to moment programmatically (the loss function), such as when there is no clear way to know how great of an impact an action will have on achieving the final target reward (such as with long term objectives - financial trading being one such example).
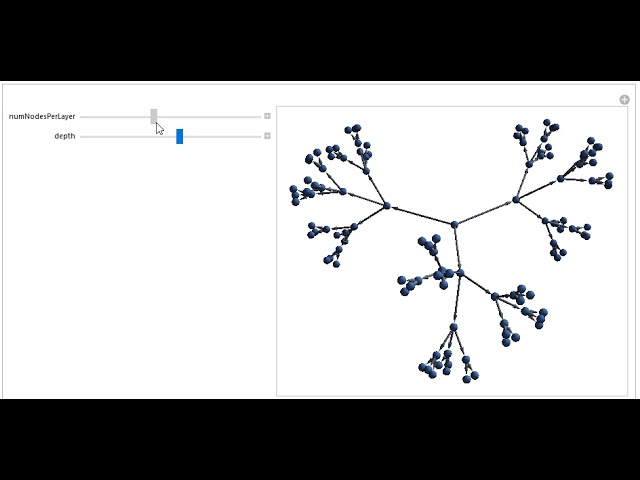
Lower Two Panes: The two panes below are videos demonstrating some of the concepts in a proposal that I made for my graduate course at Penn State in chemical nanoscience. We were asked to come up with a novel idea relating to the class, and my idea was to create a multiscale neural autoencoder-type network that successively compresses information over larger space and time scales, in an effort to predict very large scale chemical properties. Such behavior is common at the nanoscale (and can also been seen in other systems, such as cellular automata). I animated the left video by hand, which describes the general principles of the algorithm (the video contains some mildly flashing lights) and, for the video on the right, I made a program that allows one to visualize the general structure of the computational grids described in my paper. The program also allows one to modify the parameters to generate different grid shapes.
The paper is available for viewing, by clicking the following PDF button:

Genetic Reinforcement Learning

09_MY STYLE
Here, on the right and left, are videos taken from a grant proposal competition in which I participated with my group, in Dr. Deb Kelly's class in Cell and Molecular Bioengineering. We ended up winning the competition against the other students for our grant proposal on using neural networks to simulate protein folding. The video to the right is a simple visualization I made to demonstrate one aspect of the network - where the blue sphere represents the scope of attention of the neural network, relative to a single amino acid group. You may view the paper by clicking the PDF button below: